Usage¶
Valid formats¶
BitClust inherits valid trajectory and topology formats from MDTraj.
Valid trajectory extensions are: .dcd, .dtr, .hdf5, .xyz, .binpos,
.netcdf, .prmtop, .lh5, .pdb, .trr, .xtc, .xml,
.arc, .lammpstrj and .hoomdxml.
If trajectory format does not include topological information, user must pass a
path to a topology file. Valid topology extensions are: .pdb, .pdb.gz,
.h5, .lh5, .prmtop, .parm7, .prm7, .psf, .mol2,
.hoomdxml, .gro, .arc and .hdf5.
Default usage¶
Once BitClust installed, you can have access to the program’s help, which contains short descriptions of available arguments, by running
$ bitclust -h
Only one argument is always mandatory, -traj, which specifies the path to the
trajectory file. If the trajectory format does not provide topological information of
the system, it must be supplied from a topology file through the
-top argument. All other arguments are always optional and if not explicitly
specified they will take their default values commented below.
A minimal run like
$ bitclust -top tau_6K.pdb -traj tau_6K.dcd
loads the tau_6K.dcd trajectory into tau_6K.pdb coordinates (both present at
current working directory as not path was provided) and performs a clustering job using Daura’s algorithm
with a cutoff of 1A (-cutoff 1) on the whole trajectory.
Arguments -first, -last and -stride can be used to select an interval
(they default to 0, last frame and 1 respectively).
Default atom selection corresponds to all atoms (-sel all). BitClust will
retrieve all clusters with at least 2 frames (-size 2).
Frame 0 will be used as reference (-ref 0) to make an RMSD graph. All produced
output will be saved in the current working directory (-odir .).
Default outputs¶
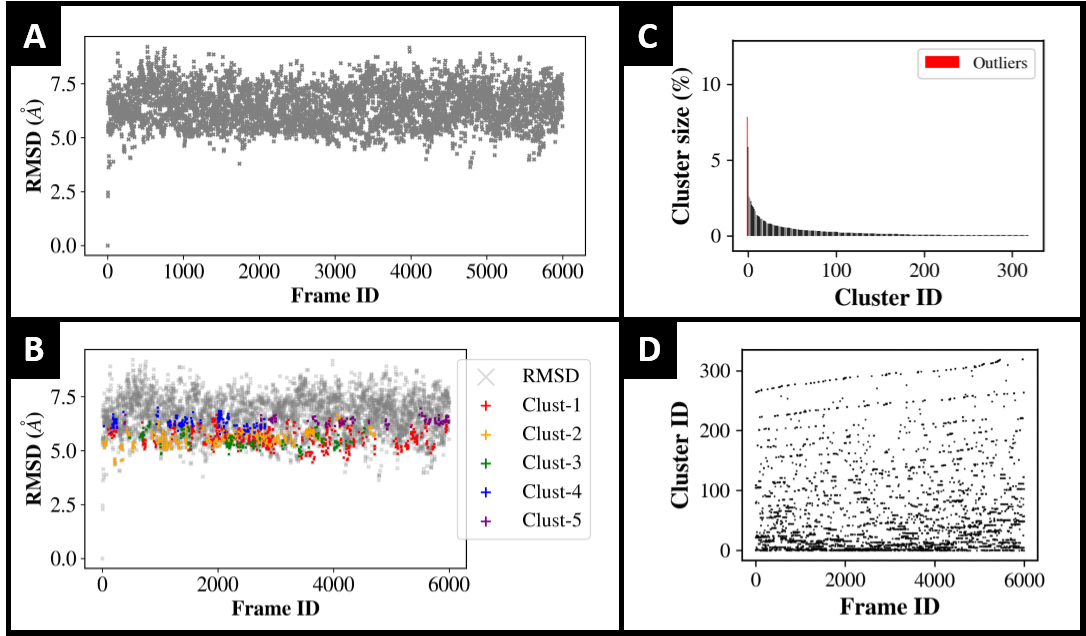
BitClust outputs basic graphics for fast inspection of the clustering job results (see figure below). All these graphs and others can be constructed from two generated text files: clusters_statistics.txt and frames_statistics.txt.
The first one contains as columns every cluster ID` (starting from 0,
-1 corresponding to unclustered frames), their size, the percent this
size represents from the total of frames and the center frame of every cluster.
The second file contains as columns every frame ID (starting from 0),
the cluster ID where every frame belongs to and the RMSD value of every
frame respect to the specified reference (default reference is frame 0).
BitClust’s basic graph outputs
Figure A: RMSD of all frames in trajectory versus reference frame passed to argument -ref. Useful for fast visualization of trajectory’s geometrical dispersion.
Figure B: Superposition of first five clusters onto Figure A. Useful for fast visualization of most populated clusters location along the trajectory.
Figure C: Clusters (including outliers in red) size. Useful for inspection of clusters relative population.
Figure D: Cluster lines. Useful to qualitatively assessment on temporal distribution of clusters.
Selection syntax¶
BitClust inherits atom selection syntax from MDTraj which is similar to that
in VMD. We reproduce below some of the MDTraj examples. Note that in BitClust
all keywords (or their synonyms) string are passed directly to -sel argument
as it is illustrated in the Usage Examples section. For more details on possible
syntax, please refer to MDTraj original documentation.
MDTraj recognizes the following keywords.
| Keyword | Synonyms | Type | Description |
all |
everything |
bool |
Matches everything |
none |
nothing |
bool |
Matches nothing |
backbone |
is_backbone |
bool |
Whether atom is in the backbone of a protein residue |
sidechain |
is_sidechain |
bool |
Whether atom is in the sidechain of a protein residue |
protein |
is_protein |
bool |
Whether atom is part of a protein residue |
water |
is_water, waters |
bool |
Whether atom is part of a water residue |
name |
str |
Atom name | |
index |
int |
Atom index (0-based) | |
type |
element, symbol |
str |
1 or 2-letter chemical symbols from the periodic table |
mass |
float |
Element atomic mass (daltons) | |
residue |
resSeq |
int |
Residue Sequence record (generally 1-based, but depends on topology) |
resid |
resi |
int |
Residue index (0-based) |
resname |
resn |
str |
Residue name |
rescode |
code, resc` |
str |
1-letter residue code |
chainid |
int |
Chain index (0-based) |
Operators¶
Standard boolean operations (and, or, and not) as well as their
C-style aliases (&&, ||, !) are supported. The expected logical
operators (<, <=, ==, !=, >=, >) are also available, as
along with their FORTRAN-style synonyms (lt, le, eq, ne,
ge, gt).
Range queries¶
Range queries are also supported. The range condition is an expression of
the form <expression> <low> to <high>, which resolves to <low> <=
<expression> <= <high>. For example
# The following queries are equivalent
-sel "resid 10 to 30"
-sel "(10 <= resid) and (resid <= 30)"
Usage examples¶
Next, you will find some usage examples of BitClust.
# An interval (1000 frames) of tau_6K.pdb trajectory (no topology file is needed)
# will be clustered with default values for all other arguments (see help section).
$ bitclust -traj tau_6K.pdb -first 0 -last 100000 -stride 100
# Clustering all atoms but hydrogen´ ones.
$ bitclust -top tau_6K.pdb -traj tau_6K.dcd -sel "\"name =~ '[^H.*]'\""
# Backbone atoms of trajectory tau_6K.dcd will be clustered using a cutoff of 4 A.
# Retrieved clusters will have at least 15 frames and output RMSD graphs will use
# frame 2580 (counting from 0) as a reference structure.
$ bitclust -top tau_6K.pdb -traj tau_6K.dcd -sel "backbone" -cutoff 4 -minsize 15 -ref 2580
# Default run saving results to local/test/run1 (relative path to current working directory)
$ bitclust -traj tau_6K.pdb -odir "local/test/run1"